The spectrum of chemoattractants and ligand sensing in Rhizobia
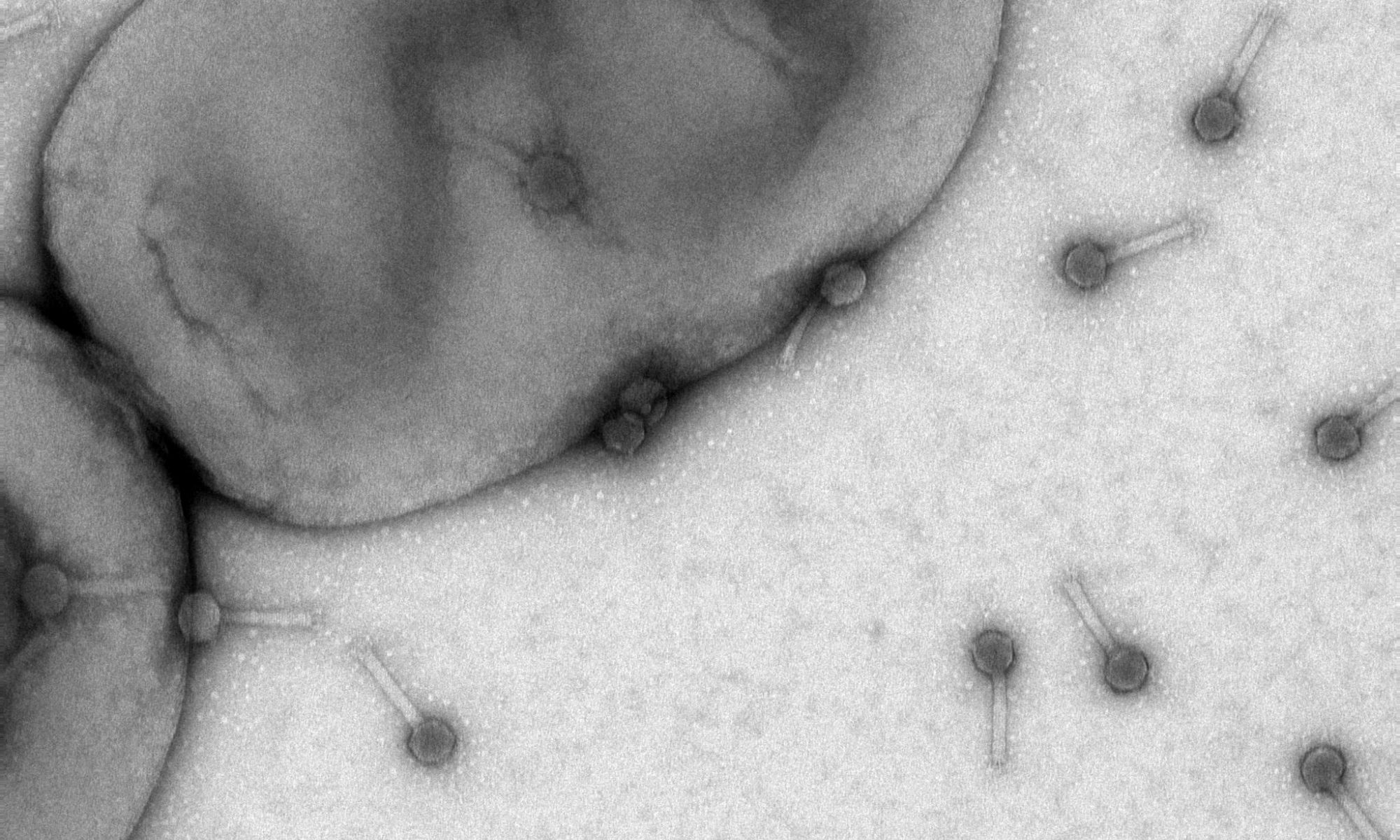
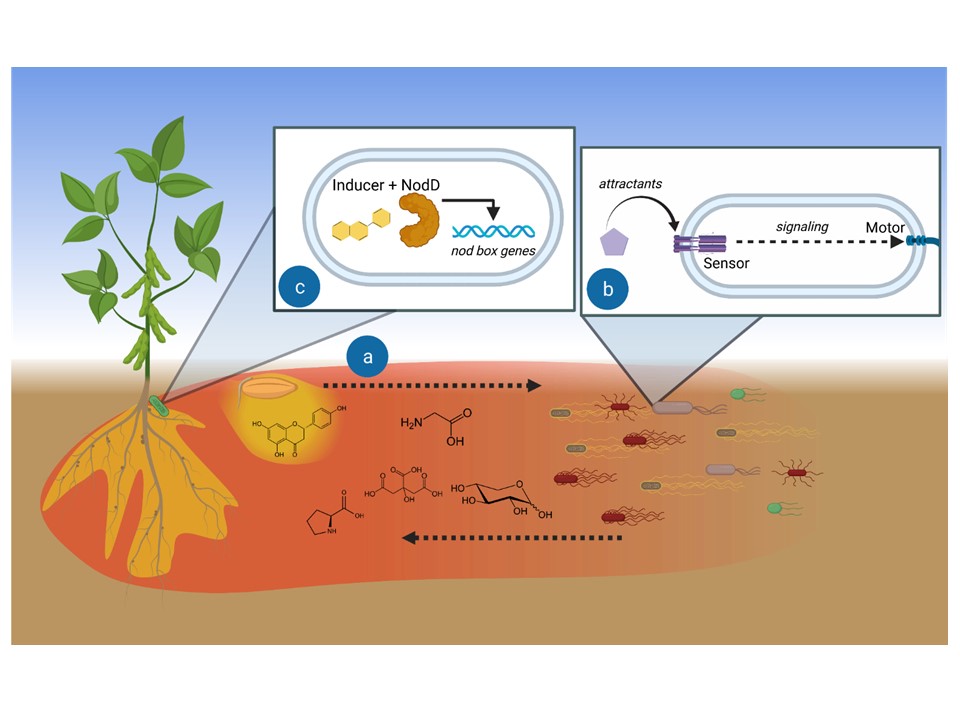
Using the alfalfa-Sinorhizobium meliloti interaction as a model system, we aim to elucidate the molecular mechanisms that govern legume-rhizobia communications. In S. meliloti, a varying soil milieu, metabolic diversity, and specific adaptations to host signals have led to the evolution of complex chemosensory system. Eight chemoreceptors play a pivotal role in directing the bacterium toward nutrients sources and its plant host. We uncovered the central role of the chemoreceptor McpU in host-plant sensing. McpU is a direct sensor of most amino acids, all of which are exuded in millimolar concentrations by germinating alfalfa seeds. Recently, we discovered that S. meliloti McpX constitutes the first known bacterial betaine chemoreceptor. Betaines protect plants against environmental stress, but their role in communication with bacteria is a new discovery. The overarching goal of this project is to characterize the specific adaptations of S. meliloti chemoreception to the range of host-derived attractants.
References:
Salar, Ball N.E., Baaziz H., Nix J.C., Sobe R.C., Compton K.K., Zhulin I.B., Brown A.M., Scharf B.E., Schubot F.D. (2023) The structural analysis of the periplasmic domain of Sinorhizobium meliloti chemoreceptor McpZ reveals a novel fold and suggests a complex mechanism of transmembrane signaling. Proteins: Structure, Function, and Bioinformatics.
H. Baaziz, K.K. Compton, Hildreth S.B., Helm R.F., Scharf B.E (2021) McpT, a broad range carboxylate chemoreceptor in Sinorhizobium meliloti. Journal of Bacteriology, 203(17), 1-16. (DOI: 10.1128/JB.00216-21)
K.K. Compton & Scharf B.E. Rhizobial chemoattractants, the taste and preferences of legume symbionts. Frontiers in Plant Science 12, 1-8. (DOI: 10.3389/fpls.2021.686465)
K.K. Compton, Hildreth S.B., Helm R.F., Scharf B.E. An updated perspective on Sinorhizobium meliloti chemotaxis to alfalfa flavonoids. Frontiers in Microbiology Vol. 11. (DOI:10.3389/fmicb.2020.581482)
M. Shrestha, Compton K.K., Mancl J., Webb B.A., Scharf B.E., Schubot, F.D. (2018) Crystal structure of the sensory domain of the first known bacterial betaine chemosensor: The methyl-accepting chemotaxis protein X (McpX) from Sinorhizobium meliloti. Biochemical Journal 14; 475(24), 3949-3962.
K.K. Compton, Hildreth S.B., Helm R.F., Scharf B.E. (2018) The Sinorhizobium meliloti chemoreceptor McpV senses short chain carboxylates via direct binding; Journal of Bacteriology 200(23), 1-16.
H.M. Zatakia, Arapov T.D., Meier V.M., Scharf B.E. (2018) Cellular stoichiometry of methyl-accepting chemotaxis proteins in Sinorhizobium meliloti. Journal of Bacteriology, 200(6), 1-14.
B.A. Webb, Compton, K.K., Martin del Campo J.S., Taylor D., Sobrado P., Scharf B.E. (2017) Sinorhizobium meliloti chemotaxis to multiple amino acids is mediated by chemoreceptor McpU. Molecular Plant Microbe Interactions 30 (10), 770-777. (DOI: 10.1094/MPMI-04-17-0096-R)
B.A. Webb, Compton K.K., Saldaña R.C., Arapov T., Ray W.K., Helm R.F., Scharf B.E. (2017) Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor McpX. Molecular Microbiology 103(2), 333-346. (DOI: 10.1111/mmi.13561)
B.A. Webb, Helm R.F., Scharf B.E. (2016) Contribution of individual chemoreceptors to Sinorhizobium meliloti chemotaxis towards amino acids of host and non-host seed exudates. Molecular Plant Microbe Interactions 29(3), 231-239. (DOI:10.1094/MPMI-12-15-0264-R )
B. Webb, Hildreth S., Helm R.F., and Scharf B.E. (2014) The Sinorhizobium meliloti chemoreceptor McpU directly binds proline to mediate chemotaxis towards host plant exudates. Applied & Environmental Microbiology 80(11), 3404-3415. (DOI:10.1128/AEM.00115-14 )
V. M. Meier and B. E. Scharf (2009) Cellular localization of predicted transmembrane and soluble chemoreceptors in Sinorhizobium meliloti. Journal of Bacteriology 191, 5724-33. (DOI:10.1128/JB.01286-08 )
V. M. Meier, Muschler, P., and Scharf B. E. (2007) Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti. Journal of Bacteriology 189, 1816-1826. (DOI:10.1128/JB.00883-06 )