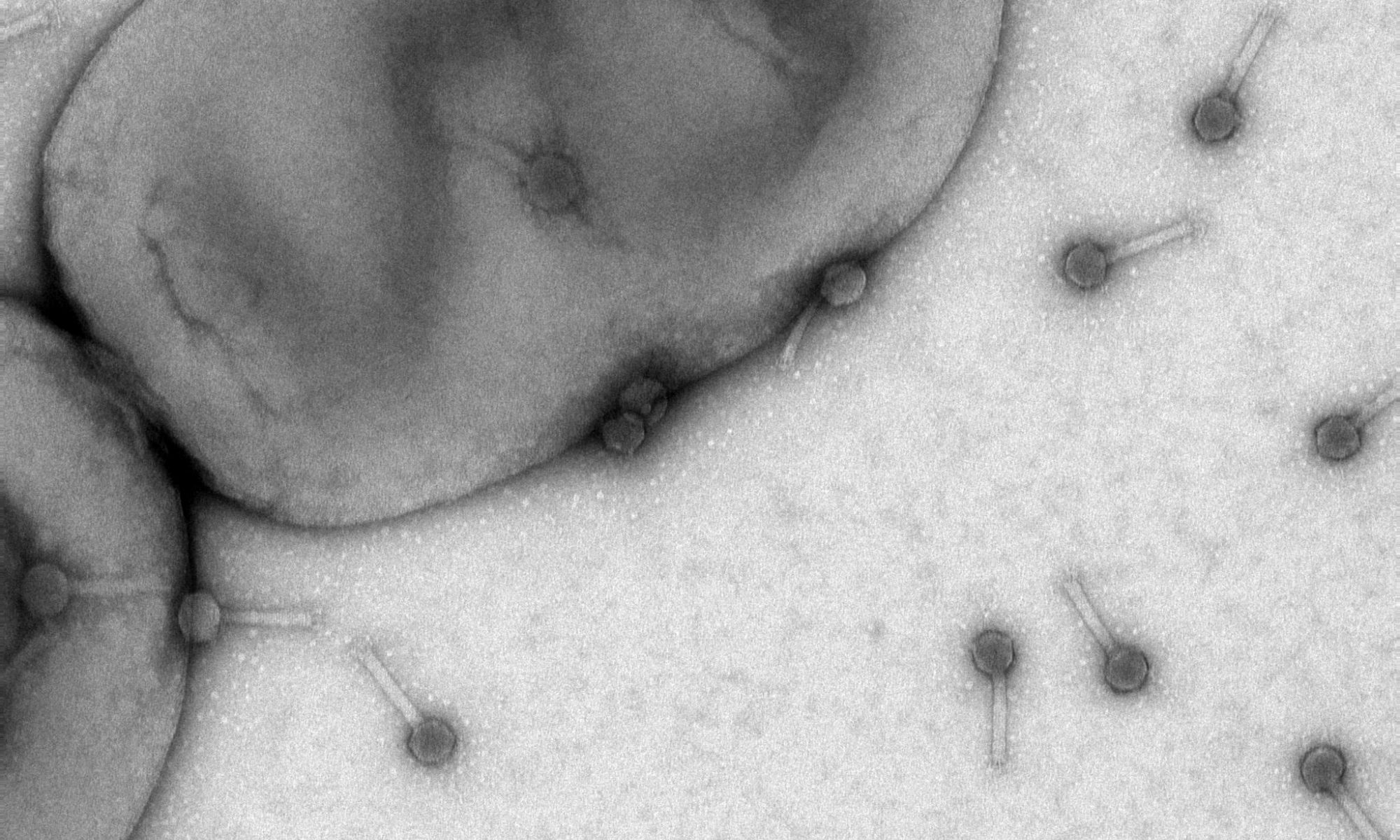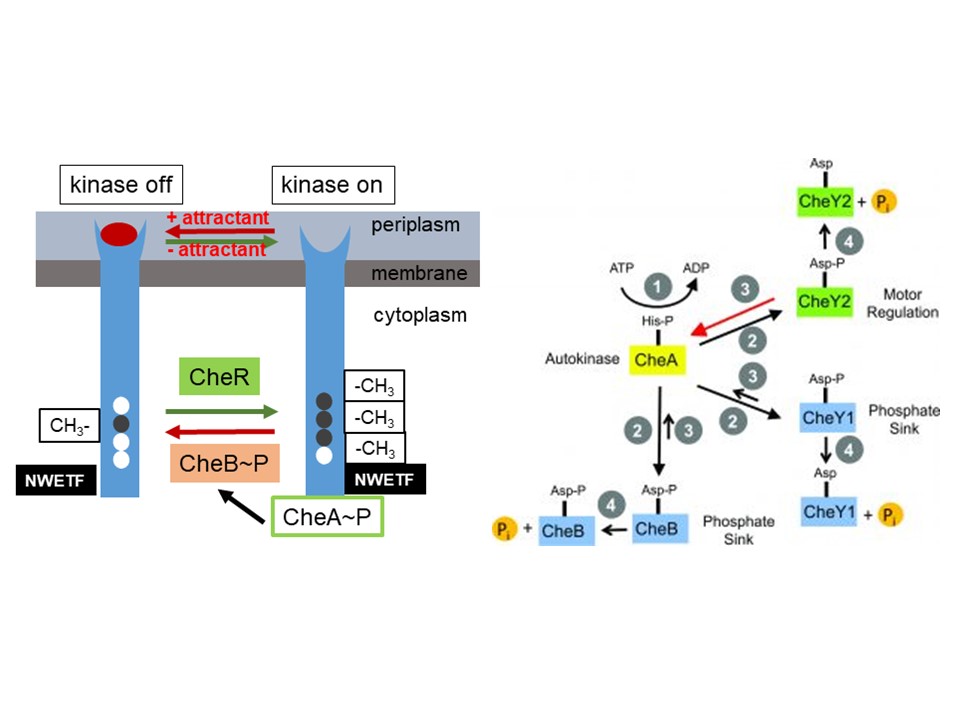
Novel aspects in bacterial signal processing, adaptation, and termination
Motility and chemotaxis are important determinants of host recognition and association by many pathogenic and beneficial bacteria. Sinorhizobium meliloti and other rhizobia have specifically adapted these systems for survival in the soil and optimal interaction with their hosts. Our overarching goal is to analyze the novel aspects of this early and important step of symbiosis that governs communication between rhizobia and their hosts. At this point, it is not understood how signal adaptation and termination are regulated. The ultimate goal of this project is to determine the reaction mechanisms and cellular stoichiometries of the key players in chemotaxis, which will allow more detailed experimental approaches and theoretical modeling of bacterial chemotaxis in the soil and during host-interaction. This knowledge majorly impacts agricultural and environmental issues by enhancing our understanding of the cues that plant symbionts and pathogens use to interact with their hosts.
References:
A. Agbekudzi & Scharf B.E. (2024) “Chemoreceptors in Sinorhizobium meliloti require minimal pentapeptide tethers to provide adaptational assistance” Molecular Microbiology, in press.
T.D. Arapov, Kim J., Cronin R.M., Pahima M., Scharf B.E. (2020) Programmed proteolysis of chemotaxis proteins in Sinorhizobium meliloti: Features in the C-terminal region control McpU degradation. Journal of Bacteriology, 202(17), 1-15. [Selected for the Spotlight Section of the Journal]
T.D. Arapov, Castañeda Saldaña R., Sebastian A.L., Ray W.K., Helm R.F., Scharf B.E. (2020) Cellular stoichiometry of chemotaxis proteins in Sinorhizobium meliloti. Journal of Bacteriology; 202(14), 141-120. (DOI: 10.1128/JB.00141-20)
T.D. Arapov, Kim J., Cronin R.M., Pahima M., Scharf B.E. (2020) Programmed proteolysis of chemotaxis proteins in Sinorhizobium meliloti: Features in the C-terminal region control McpU degradation. Journal of Bacteriology. 202(17), 1-15. [Selected for the Spotlight Section of the Journal] (
D. Dogra, Purschke F.G., Wagner V., Haslbeck M., Kriehuber T., Hughes J.G., Van Tassell M.L., Gilbert C., Niemeyer M., Ray W.K., Helm R.F., Scharf B.E. (2012) Sinorhizobium meliloti CheA complexed with CheS exhibits enhanced binding to CheY1 resulting in accelerated CheY1-P dephosphorylation. Journal of Bacteriology 194, 1075-1087. [cited in faculty of 1000] (DOI:10.1128/JB.06505-11)
D. Wibberg, Blom J., Winkler A., Albersmeier A., Pühler A., Scharf B.E. (2013) Draft genome sequence of Sinorhizobium meliloti RU11/001, a model organism for flagellum structure, motility and chemotaxis. Journal of Biotechnology 4, 731-733. (DOI:10.1016/j.jbiotec.2013.10.015)
H. Riepl, Maurer T., Kalbitzer H.R., Meier V.M., Schmitt R., Scharf B. (2008) Interaction of CheY2 and CheY2-P with the cognate CheA kinase in the chemosensory-signalling chain of Sinorhizobium meliloti. Molecular Microbiology 69, 1373-1384. (DOI:10.1111/j.1365-2958.2008.06342.x)
H. Riepl, Scharf B., Schmitt R., Maurer T., Kalbitzer H.R. (2004) Solution structures of the inactive and BeF3-activated response regulator CheY2. 338, 287-297. [cited in faculty of 1000] (DOI:10.1016/j.jmb.2004.02.054 )
R.C. Sobe, Gilbert C., Vo L., Alexandre G., Scharf B.E. (2022) FliL and its paralog MotF have distinct roles in the stator activity of the Sinorhizobium meliloti flagellar motor. Molecular Microbiology 118(3), 223-243.
- M.A.B. Kreutzberger, Sonani R.R., Liu J., Chatterjee S., Wang F., Sebastian A.L., Biswas P., Ewing C., Zheng W., Poly F., Frankel G., Luisi B.F., Calladine C.R., Krupovic M., Scharf B.E., Egelman E.H. (2022) Convergent evolution in the supercoiling of prokaryotic flagellar filaments. Cell 15;185(19):3487-3500.
- S. Salar, Ball N.E., Baaziz H., Nix J.C., Sobe R.C., Compton K.K., Zhulin I.B., Brown A.M., Scharf B.E., Schubot F.D. (2023) The structural analysis of the periplasmic domain of Sinorhizobium meliloti chemoreceptor McpZ reveals a novel fold and suggests a complex mechanism of transmembrane signaling. Proteins: Structure, Function, and Bioinformatics.; 91(10):1394-1406.
- N.C. Esteves, Bigham D.N., Scharf B.E. (2023) Phages on filaments: A genetic screen elucidates the complex interactions between Salmonella enterica flagellin and bacteriophage Chi, PLOS Pathogens; 19(8).
- R.R. Sonani, Esteves N.C., Horton A.A., Kelly R.J., Sebastian A.L., Wang F., Kreutzberger M.A.B., Leiman P.G.*, Scharf B.E.*, Egelman E.H.* (2023) “Neck and capsid architecture of the robust Agrobacterium phage Milano” Springer Nature Communications Biology; 6(1):921.
* Co-corresponding authors.
- R.R. Sonani, Palmer L.K., Esteves N.C., Horton A.A., Sebastian A.L., Kelly R.J., Wang F., Kreutzberger M.A.B., Russell W.K., Petr G. Leiman P.G.*, Scharf B.E.*, Egelman E.H.* (2024) “An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano” Springer Nature Communications Biology; 15(1):756.
* Co-corresponding authors.
- R.C. Sobe & Scharf B.E. (2024) “The swimming defect caused by absence of the transcriptional regulator LdtR in Sinorhizobium meliloti is restored by mutations in the motility genes motA and motS” Molecular Microbiology; 121(5):954-970.
- R.R. Sonani, Esteves E.C., Scharf B.E.*, Egelman E.H.* (2024) “Cryo-EM structure of flagellotropic bacteriophage Chi” Structure, 024 Apr 3:S0969-2126(24)00093-5. doi: 10.1016/j.str.2024.03.011. Online ahead of print.
* Co-corresponding authors.
- A. Agbekudzi & Scharf B.E. (2024) “Chemoreceptors in Sinorhizobium meliloti require minimal pentapeptide tethers to provide adaptational assistance” Molecular Microbiology, in press.
T.D. Arapov, Castañeda Saldaña R., Sebastian A.L., Ray W.K., Helm R.F., Scharf B.E. (2020) Cellular stoichiometry of chemotaxis proteins in Sinorhizobium meliloti. Journal of Bacteriology; 202(14), 141-120. (DOI: 10.1128/JB.00141-20)
T.D. Arapov, Kim J., Cronin R.M., Pahima M., Scharf B.E. (2020) Programmed proteolysis of chemotaxis proteins in Sinorhizobium meliloti: Features in the C-terminal region control McpU degradation. Journal of Bacteriology. 202(17), 1-15. [Selected for the Spotlight Section of the Journal] (
D. Dogra, Purschke F.G., Wagner V., Haslbeck M., Kriehuber T., Hughes J.G., Van Tassell M.L., Gilbert C., Niemeyer M., Ray W.K., Helm R.F., Scharf B.E. (2012) Sinorhizobium meliloti CheA complexed with CheS exhibits enhanced binding to CheY1 resulting in accelerated CheY1-P dephosphorylation. Journal of Bacteriology 194, 1075-1087. [cited in faculty of 1000] (DOI:10.1128/JB.06505-11)
D. Wibberg, Blom J., Winkler A., Albersmeier A., Pühler A., Scharf B.E. (2013) Draft genome sequence of Sinorhizobium meliloti RU11/001, a model organism for flagellum structure, motility and chemotaxis. Journal of Biotechnology 4, 731-733. (DOI:10.1016/j.jbiotec.2013.10.015)
H. Riepl, Maurer T., Kalbitzer H.R., Meier V.M., Schmitt R., Scharf B. (2008) Interaction of CheY2 and CheY2-P with the cognate CheA kinase in the chemosensory-signalling chain of Sinorhizobium meliloti. Molecular Microbiology 69, 1373-1384. (DOI:10.1111/j.1365-2958.2008.06342.x)
H. Riepl, Scharf B., Schmitt R., Maurer T., Kalbitzer H.R. (2004) Solution structures of the inactive and BeF3-activated response regulator CheY2. 338, 287-297. [cited in faculty of 1000] (DOI:10.1016/j.jmb.2004.02.054 )